Workflow modelling and enactment for polymorph simulation
The UCL Theoretical Chemistry group led by Professor Sarah L. Price has a long track record in polymorph simulation. This interest is driven by the need for understanding the different possible crystal structures that a complex molecule may adopt and has important applications in drug and formulation design, understanding the behaviour of pigments in paint and the engineering of complex materials. In the EPSRC funded e-Materials project, the group ported their approach for polymorph simulation to a grid setting.
Early on in that effort it became clear that the workflows that searches carry out needed to be flexibly amended and the group experimented with BPEL for that purpose.
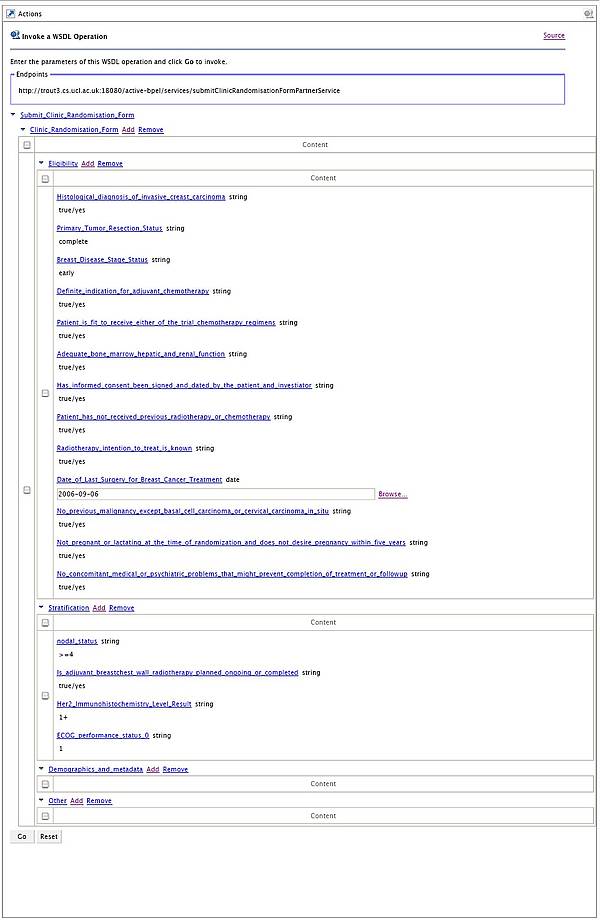
Prof. Price’s team now uses the OMII-BPEL environment, GridSAM and the entire OMII stack to control the parallel execution of up to 8000 jobs on the UCL Condor pool during any one search. The figure below shows the BPEL process that controls job-submissions to the UCL Condor by invoking web services provided by GridSAM.
This process is one of the leaves in a complex hierarchy of processes. Such hierarchical composition is enabled, as each BPEL process is also a web service. In order to achieve significant speed-up of the simulation on a grid infrastructure hundreds and thousands of these submission processes can be handled by the OMII-BPEL run-time concurrently. In grid settings nodes also occasionally fail or become unreachable and BPEL compensation handling primitives support recovery from these failures, typically through re-submission to other nodes. The environment persists process state in PostgresQL, the OMII database in order to survive accidental or scheduled termination of the BPEL run-time, a crucial feature to support long-running workflows. The OMII-BPEL environment is fully integrated with the OMII stack. As a result the execution of BPEL processes is secured through WS-Security primitives, a crucial feature required to protect the IP of drug discovery processes in a grid setting.
Through this grid-enabled approach, Prof. Price’s group has brought down the time for obtaining simulation results from up to three months (on a dual CPU Silicon graphics machine) to mere 4 hours on the 1,200+ node UCL Condor pool and at the same time increased the precision of the searches. This dramatic improvement has changed the way chemists perform simulations and enabled the simulation of more complex molecules. Prof. Price’s group is now able to perform exploratory and low precision searches interactively. The provision of this environment has enabled a number of scientific breakthroughs. Using the approach Prof. Price’s group was able to predict a new polymorph of the drug Aspirin, which was later experimentally confirmed. The BPEL environment was used to successfully confirm the structure of a new polymorph of a peptide-coupling agent. Most publicity, including an article in Nature News has been generated by the successful prediction of the structure of the conformational polymorph IV of the nootropic agent Piracetam, which was recently discovered by re-crystallisation under pressure.
For further information see:
- <link http://dx.doi.org/10.1007/s10723-005-9015-3>Emmerich, W.; Butchart, B.; Chen, L.; Wassermann, B.; Price, S.; J Grid Comp 2005, 3(3-4), 283-304</link>
- Ouvrard, C.; Price, S. L. Cryst. Growth Des. 2004, 4, 1119.
- Nowell, H.; Frampton, C. S.; Waite, J.; Price, S. L. Acta Crystallogr. Sect. B-Struct. Sci. 2006. In press.
- <link http://www.nature.com/news/2005/050919/full/050919-7.html>Nature News, 5/9/2005</link>
- Nowell, H.; Price, S. L. Acta Crystallogr. Sect. B-Struct. Sci. 2005, 61, 558.
Management of clinical workflows in cancer research
The EPSRC and MRC funded CancerGrid project will deliver modular, distributed software solutions for the key problems of clinical cancer informatics: clinical trial patient entry, randomisation and follow-up; storage and analysis of complex datasets; linking trial and epidemiology data with profiling information. It will facilitate trials management, and future collaboration across international boundaries. CancerGrid has adopted a .NET-based web-services approach and these web services need to be combined in a flexible manner and these combinations then form new web services themselves. In 2005, CancerGrid has adopted BPEL as their workflow language and the ActiveBPEL engine contained in the OMII-BPEL environment for modelling, enacting and monitoring a large number of clinical workflows for cancer research. CancerGrid are developing web services to handle patient records in cancer research.
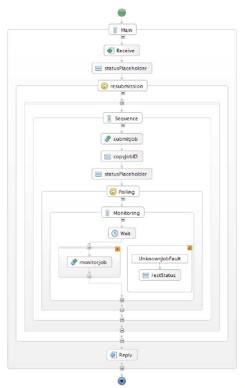
The figure below shows an early CancerGrid workflow that combines two web services. The first service validates whether the details provided about a patient are complete and valid and whether the patient is eligible to participate in a trial. If this is the case the workflow then invokes a service that determines the most appropriate treatment type for the service. The figure to the right is a print-out of the BPEL representation that was developed with the Eclipse-based BPEL Designer that UCL develop jointly with Oracle, IBM and JBoss and that is integrated into the OMII-BPEL environment. The workflow was determined, validated and deployed into the ActiveBPEL run-time environment within 15 minutes. As an Eclipse-environment, BPEL Designer provides support for version and configuration management of workflows, validation of workflows prior to deployment, support to edit, browse and query service interface definitions in WSDL and type definitions in XML Schema and many other useful features.
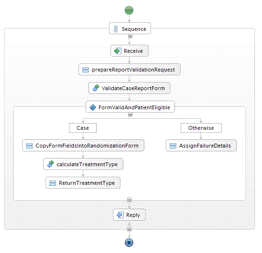
Following deployment of a BPEL process, BPEL Designer can generate a Web Service Explorer that serves as a web-based user interface and allows users to invoke the web service implemented by the BPEL process. This is achieved by a single BPEL Designer command. The figure below shows the Web Service Explorer for the clinical workflow shown above.